Epidemic spreading under mutually independent intra- and inter- host pathogen evolution – Nature Communications
Stefano Boccaletti, researcher @ ISC – Firenze, coauthored an interesting work discussing how evolving pathogens impact the reproduction number and macroscopic dynamics of spreading processes. The article is also featured in the Applied Physics and Mathematics Focus page selecting “the most interesting papers published in Nature Communications in the interdisciplinary areas where diverse approaches at the boundaries of physics, mathematics, materials science and engineering take place to create new research opportunities”
Epidemic spreading under mutually independent intra- and inter-host pathogen evolution, X. Zhang, Z. Ruan, M. Zheng, S. Boccaletti and B. Barzel Nat. Commun. 13, 6218 (2022).
Abstract
The dynamics of epidemic spreading is often reduced to the single control parameter R0 (reproduction-rate), whose value, above or below unity, determines the state of the contagion. If, however, the pathogen evolves as it spreads, R0 may change over time, potentially leading to a mutation-driven spread, in which an initially sub-pandemic pathogen undergoes a breakthrough mutation. To predict the boundaries of this pandemic phase, we introduce here a modeling framework to couple the inter-host network spreading patterns with the intra-host evolutionary dynamics. We find that even in the extreme case when these two process are driven by mutually independent selection forces, mutations can still fundamentally alter the pandemic phase-diagram. The pandemic transitions, we show, are now shaped, not just by R0, but also by the balance between the epidemic and the evolutionary timescales. If mutations are too slow, the pathogen prevalence decays prior to the appearance of a critical mutation. On the other hand, if mutations are too rapid, the pathogen evolution becomes volatile and, once again, it fails to spread. Between these two extremes, however, we identify a broad range of conditions in which an initially sub-pandemic pathogen can breakthrough to gain widespread prevalence.
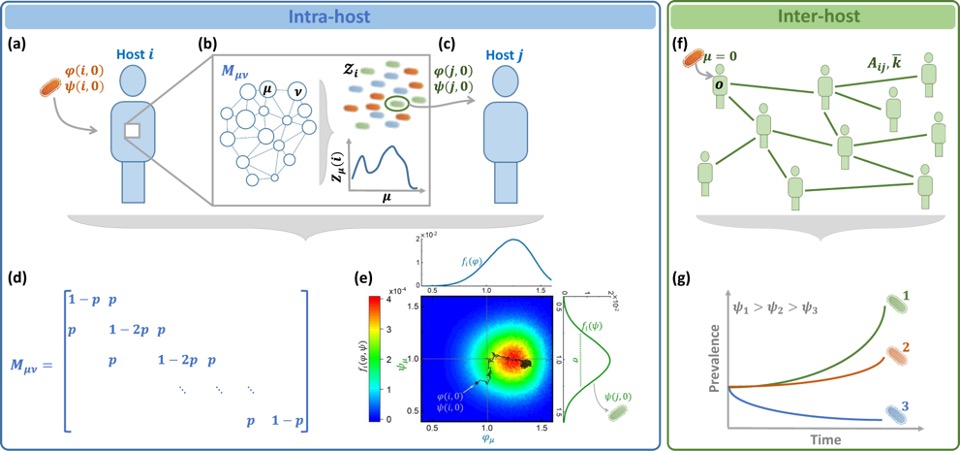
The interplay between evolutionary and epidemiological dynamics. (a) Host i contracts a pathogen (orange) with initial fitness φ(i, 0), ψ(i, 0). (b) Within i the pathogen replicates, undergoing mutation and selection. (c) Upon transmission i → j, a single (or few) individual pathogens instigate again j’s intra-host dynamics, this time staring from initial fitness φ(j, 0), ψ(j, 0). (d) The transition matrix Mμν. Mutations enable transition with probability 0 ≤ p ≤ 1 between adjacent strains. The larger is p, the less stable is the pathogen. (e) The fitness distribution fi(φ, ψ) following ρ = 25, 000 replication/mutation cycles initiated at an arbitrary φ(i, 0), ψ(i, 0). (f) The inter-host dynamics is captured by network epidemic spreading. (g) The network spreading dynamics give rise to selection for higher ψμ.

