Protein Folding
Protein Folding is a specific chemical and physical transition by which a linear sequence of aminoacids finds its functional (native) three dimensional structure. The theoretical study of protein folding represents perhaps one of the most challenging research with a marked interdisciplinary character, where biology, chemistry, physics, mathematics and computer science can fruitfully interact each other.
Our activity in this field concerns the prediction of folding mechanisms by the knowledge of tertiary structure only. This finds a strong justification on the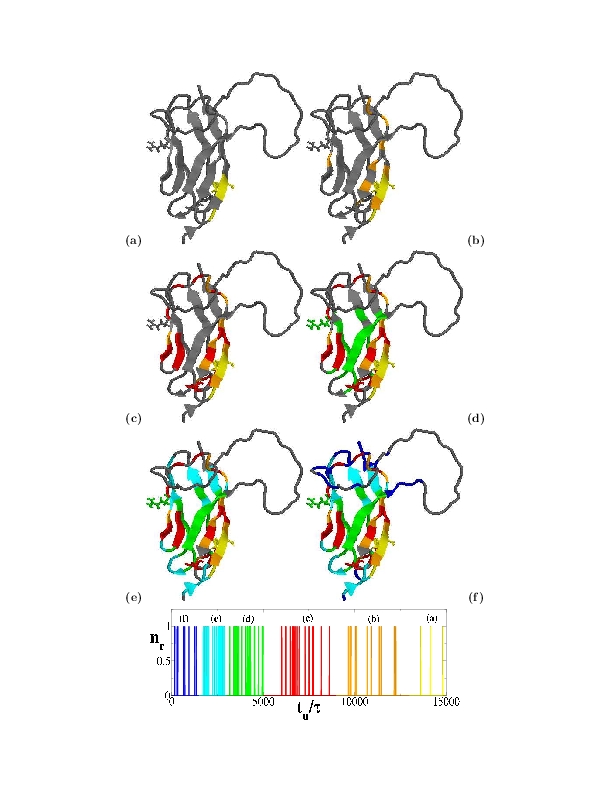 widely recognised importance that native states have in stirring the folding processes.
widely recognised importance that native states have in stirring the folding processes.
In particular, we follow the approach combining molecular dynamics simulations, coarse-grained protein models and Statistical Mechanical methods which constitutes a viable framework to tackle this issue at convenient computational costs. In this philosophy the protein is assimilated to a polymer composed of its backbone of C-alpha carbons and the interactions among amino-acids are assigned via an potential energy which takes on a minimum on the native state.
This modeling is called native-centric (or Go-like) approach as is built up around the native state structure. Typical folding pathways predicted by a native-centric model are shown in the figure, where colors indicate the different stages of native secondary structure formation.
Our interest in folding-pathways prediction by tertiary structure is also focused on potential biomedical implications with the purpose to establish and tune a computer-aided procedure to characterize pathogenicimpact of mutations.
In a series of papers, we tested the potentiality of native-centric coarse-grained modelling in investigating the folding properties of wild type and mutants of domain C5 from cardiac myosin binding protein C (MyBPC). We suggest to asses the impact of a mutation through the change in protein stability and unfolding kinetic rates and through the knowledge of folding pathways.
Kinetics simulations reveal a well defined folding pathway governed by the immunoglobulin-topology of the C domain (see figure).
The folding pathway allows a possible interpretation of the molecular impact of mutations involved in the Familial Hypertrophic Cardiomyopathy because the effect of a mutation gains relevance in the proximity to the area where folding originates. The scenario is confirmed by the kinetic pathways of test mutations evenly distributed throughout the entire C5 domain.
[1] F. Cecconi, C. Micheletti, P. Carloni and A. Maritan,“Molecular Dynamics Studies on HIV-1 Protease:
Drug Resistance and Folding Pathways” Proteins: Struc. Funct. Gen. 43, 365 (2001).
[2] F. Cecconi, C. Guardiani and R. Livi, “Testing simplified protein models of the hPin1 WW domain”
[3] C. Guardiani, F. Cecconi and R. Livi,”Computational analysis of folding and mutation properties of C5 domain from
Myosin Binding Protein C”, Proteins: Struct. Func. Bioinf. 70, 1313 (2008).
[4] C. Guardiani, F. Cecconi and R. Livi, “Stability and kinetic properties of C5-domain from Myosin Binding Protein C
and its mutants” Biophys. J. 94, 1403 (2008).
[5] F. Cecconi, C. Guardiani and R. Livi,”Analyzing pathogenic mutations of C5 domain from cardiac Myosin Binding
Protein C through MD simulations” Eur. Biophys. J. 37, 683 (2008).



