NAR: Energetic funnel facilitates facilitated diffusion
Massimo Cencini has published Energetic funnel facilitates facilitated diffusion in Nucleic Acid Research.
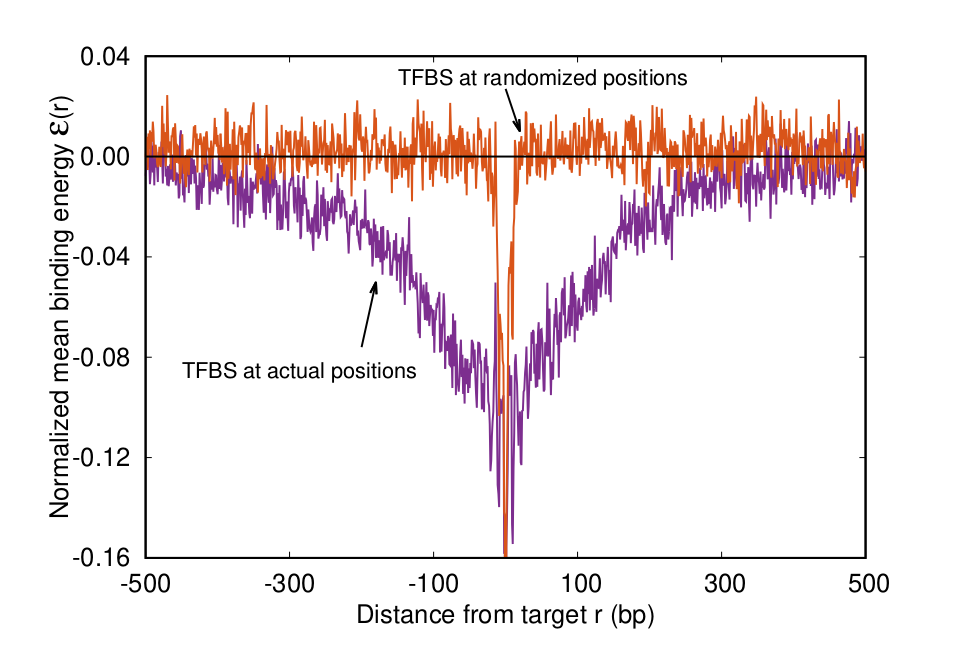
FIG. 1: Average free binding energy landscape around the bibnding sites of TFs in E. coli. The comparison with the null model obtained by placing the target sites in random genome positions reveal the funnel structure of the energy landscape.
Gene transcription is regulated by proteins – Transcription Factors (TFs) – that by binding to short target sequences are able to promote or impede the binding of RNA-Polymerase (RNAP) and, consequently, activate or repress tran-
scription. Fast and accurate control of gene expression is crucial for many biological functions, and relies on the ability of TFs to rapidly find their transcription factor binding site (TFBS) among a multitude of competing DNA sequences,
and to establish with it a stable complex. Experimental studies have shown that TFs are able to associate to their
binding sites on DNA faster than the physical limit posed by diffusion. Theoretical studies have speculated that such
high association rates can be achieved by alternating between three-dimensional diffusion and one-dimensional sliding
along the DNA chain, a mechanism-dubbed Facilitated Diffusion.
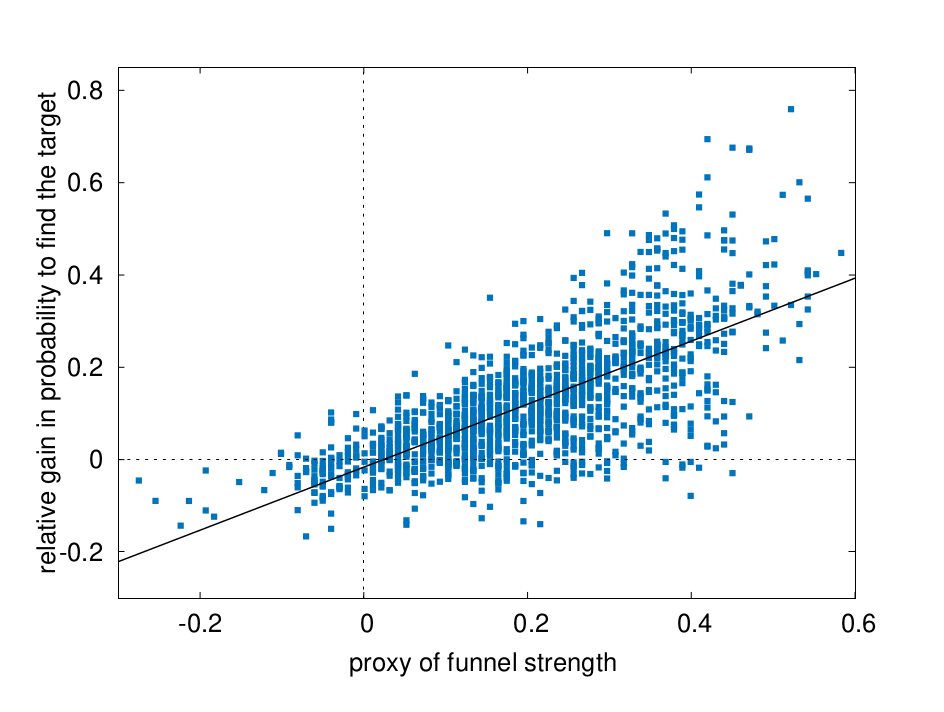
FIG. 2: This scatter plot shows the gain in probability to find the target (relative to the null model obtained by placing the target sites in random genome positions) versus the funnel strength around the 1544 target sites present in the analysed database.
In this work, by studying a collection of 1544 TF binding sites of Escherichia coli from the RegulonDB database
and 313 of Bacillus subtilis from DBTBS, it was revealed a funnel in the binding energy landscape around the target
sequences (see Fig. 1). Moreover, such a funnel was found to be linked to the presence of gradients of AT in the base
composition of the DNA region around the binding sites. Finally, by means of an extensive computational study of
a biophysical model for the sliding process along the energetic landscapes obtained from the database, it was shown
that the funnel can significantly enhance the probability of TFs to find their target sequences when sliding in their
proximity (see Fig. 2) leading to a sensitive shortening of the search time.
These results provide new insights on the mechanisms of Facilitate Diffusion that can stimulate new experiments
on transcription factors in prokaryotes. In particular, may suggest methods to enginering articificial promoters to be
used in synthetic biology.
